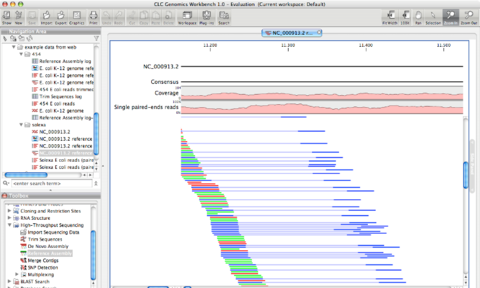
CLC Bio has released version 5.5 of its desktop software, CLC Genomics Workbench, alongside CLC Genomics Server version 4.5.
Both software packages have been updated with a range of features, especially breakthrough additions in functional genomics, now supporting a complete resequencing workflow from read mapping over variant detection to downstream analysis.
“Until today, no other tools allowed non-programmers to build resequencing workflows through a graphical user-interface.
“With this release we’re enabling biologists, clinicians, and other scientists to intuitively build workflows and visualise their datasets through all stages of the analysis - right on their desktop computers.” said VP of R&D at CLC bio, Dr. Roald Forsberg.
For example, users can build a complete analysis workflow, starting with read trimming, followed by mapping, calling and annotating variants, and ending with comparative analysis.
The workflows can be used to combine several filtering and annotation steps into one, and run in batch mode to analyse a high number of samples.
Features include:
- Workflows can be created to combine various tools from the toolbox into one single analysis step
- Tracks, the new way of storing, comparing and displaying genomics data, can easily be created and used in all resequencing tools
- Advanced variant detection, also well suited for genomes of higher ploidy
- Trio analysis comparing father-mother-child variants enables studies of inherited and de novo mutations
- Download of genomics sequence and annotation data from public databases




