The RNase H2–dependent polymerase chain reaction (rhPCR) method from Integrated DNA Technologies (IDT) is helping researchers to detect differential amplification of alternatively spliced latrophilin (Lphn) transcripts in mice.
Latrophilins are cell adhesion molecules that have been linked with synaptic function, development, and maintenance.
The three Lphn genes are very similar, which complicates the use of standard qRT-PCR to distinguish the various transcripts.
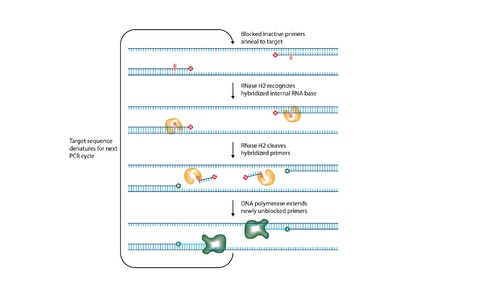
The rhPCR method developed by IDT overcomes this problem by simultaneously increasing PCR specificity and eliminating primer-dimer formation.
This is achieved by using blocked primers, which require cleavage by RNase H2 before they can be used for target amplification.
The RNase H2 enzyme is active at standard PCR temperatures and cleaves the 5’ end of a single RNA base in the blocked primer only after the primer hybridizes to its target.
Therefore, the primers are unable to form primer-dimers, and with the required high complementarity to their targets, reduce amplification of closely related sequences.
The cleaved, unblocked primers are then available for extension and replication.
In its web publication, DECODED online, IDT discusses a recent publication which extends the rhPCR method to RT-PCR by the addition of an optimised concentration of RNase H2 after an initial cDNA synthesis step.
The method successfully distinguished alternatively spliced variants, allowing Lphn expression to be quantitatively measured in various tissues.
The paper represents just one such application of IDT’s rhPCR technology, which also facilitates SNP genotyping, multiplexing, NGS library preparation, and detection of rare alleles.




