An international research team has determined the structure of a new drug target in complex with a synthetic molecule, opening up new possibilities for drug discovery.
The enzyme SHIP2 has attracted particular attention due to its role in the negative regulation of insulin signalling and is thought to be involved in type-2 diabetes, obesity and cancer.
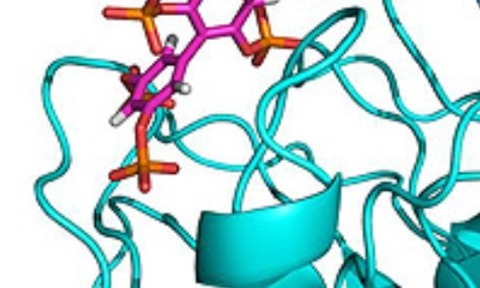
However, previous attempts to crystallise the enzyme with its natural substrate have been unsuccessful.
As part of the project, scientists at Bath University designed a synthetic inhibitor as a mimic and with their European colleagues solved the X-ray crystal structure of a key fragment of SHIP2 bound to this compound.
The scientists also undertook computational molecular dynamics on the complex, and discovered a flexible loop region of the protein that may close over the compound during binding.
The researchers hope that targeting such a closed complex could provide a new strategy for the design of small-molecule drugs against SHIP2.
Prof Barry Potter, who led the enterprise, said: “Such interdisciplinary collaboration represents a real route to early progress in drug discovery at a time when the global pharmaceutical industry is restructuring and looking more than ever towards academic-industry partnerships for early stage drug discovery, rather than in-house R&D.”
The research was a collaborative project carried out by an international group of scientists based at the Karolinska Institutet in Stockholm Sweden, Nanyang University Singapore, the Université Libre de Bruxelles in Belgium and the University of Bath.
The next step will be to design in silico related, but more drug-like, compounds that might bind to the closed complex of the SHIP2 enzyme. The researchers hope that others will use their work as a starting point to design such novel drug candidates.

